SINGLE CELL RNA SEQUENCING KIT
Single Cell Gene Expression for All
Enables single-cell RNA sequencing across dozens of samples with recovery of up to 500,000 cells or nuclei.
Description
The ScaleBio Single Cell RNA Sequencing Kit enables the production of high-quality single-cell transcriptomic libraries. Our upfront cell fixation step allows you to take a flexible approach to sample collection, whether from multiple sites or multiple timepoints. This instrument-free, two-day workflow allows you to profile up to 125,000 cells or nuclei while multiplexing up to 96 samples in a single experiment.
Specifications
Sample Compatibility
Fixation-compatible workflow, for both cells and nuclei, with storage up to 6 months.
Cell Throughput
Process 125,000 cells or nuclei per run
Sample Multiplexing
Up to 96 samples per run
Unique Combinations
Generate >3.5 million barcode combinations with three levels of indexing, while maintaining a <5% doublet rate
Single Cell RNA Sequencing workflow
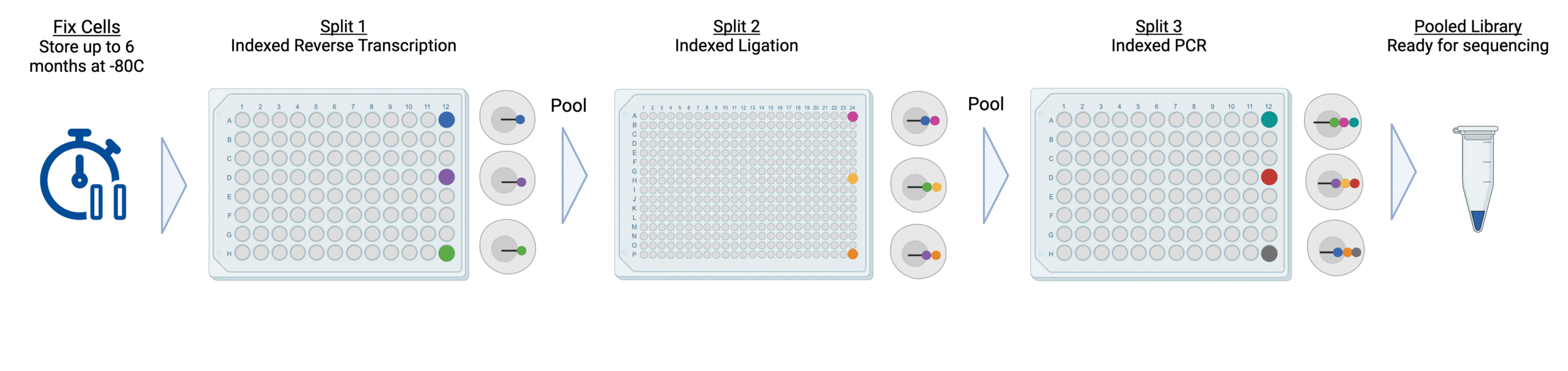
Single-cell or nuclei suspensions are prepared and fixed using the ScaleBio Sample Fixation Kit. Samples can be used immediately or stored for up to 6 months at -80°C. Cells or nuclei are then barcoded through three split-pool indexing rounds to produce a sequence-ready library, with numerous safe stopping points to accommodate your schedule. Data is analyzed with the ScaleBio Seq Suite RNA pipeline.
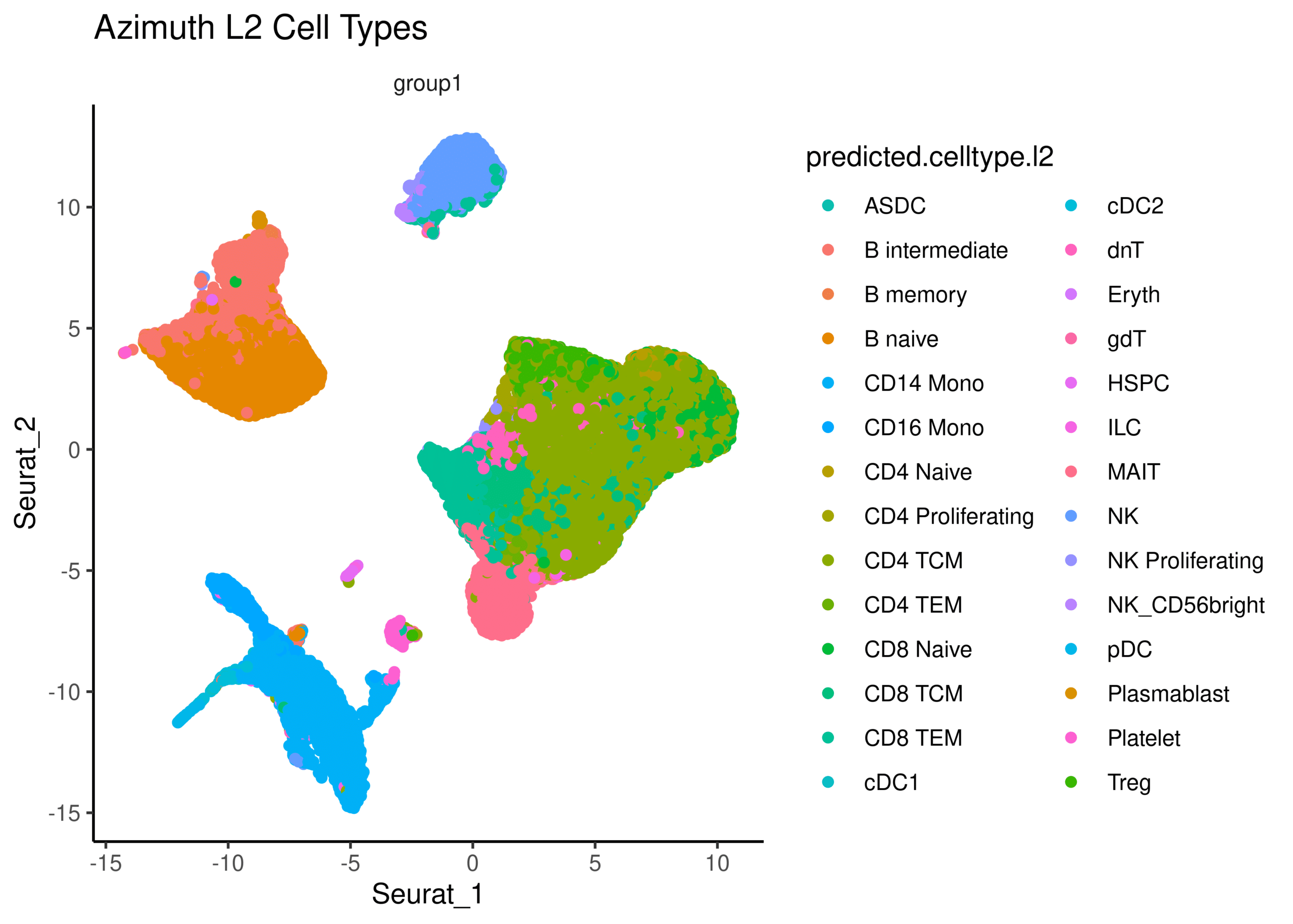
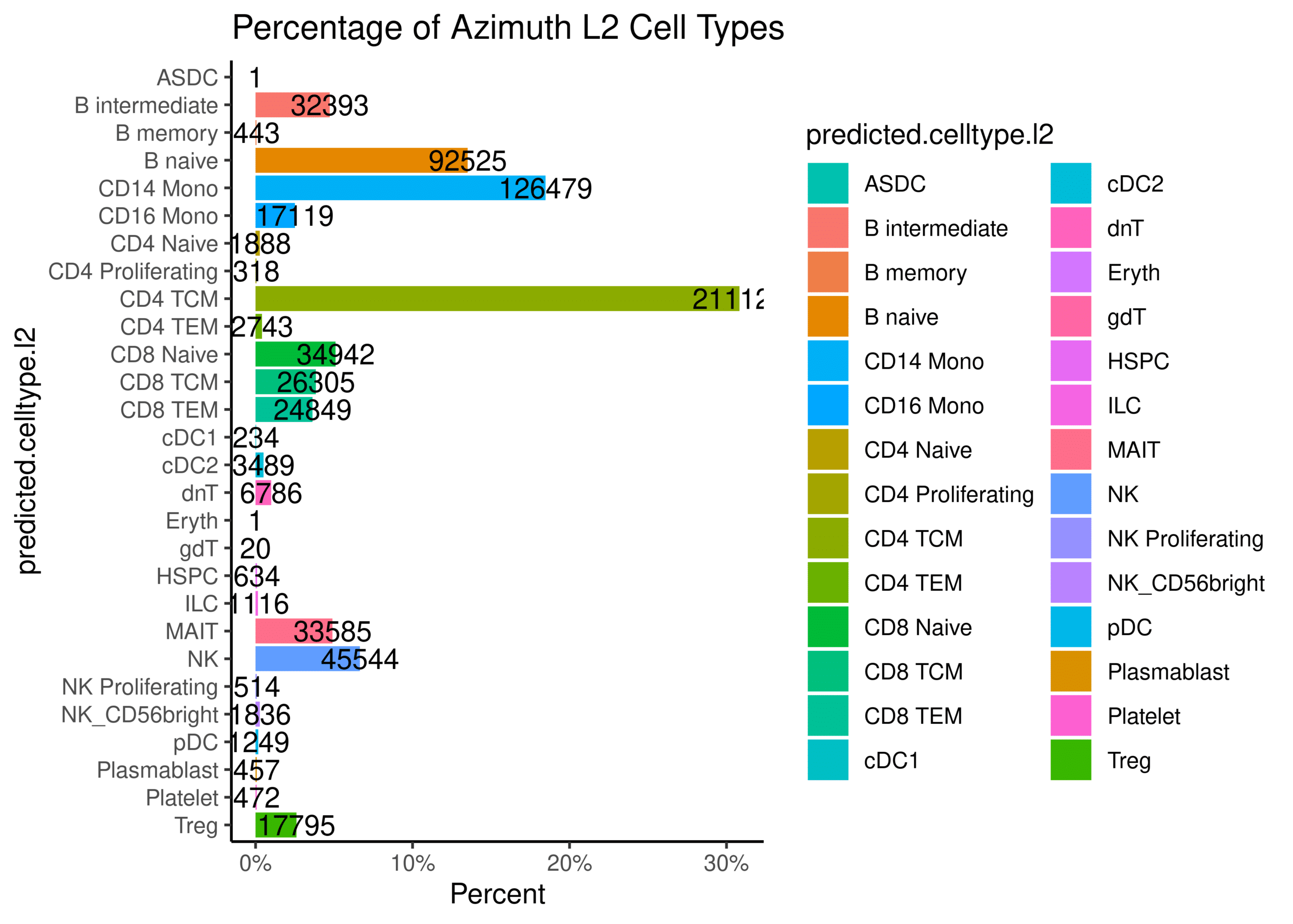
PBMC data reveals expected sample heterogeneity
Over 500,000 PBMCs were profiled to explore cellular heterogeneity within a complex sample type. The UMAP clustering on a subset of data demonstrates the cell type and cell state heterogeneity of the sample, all easily identified with ScaleBio's sensitive single-cell combinatorial indexing chemistry.
Featured publications View All
Frequently asked questions
Together with our customers, we have demonstrated performance of our Single Cell RNA Sequencing Kit across a broad range of cell types, including but not limited to: tissue-dissociated cells, nuclei isolated from frozen and fresh tissues, PBMCs, and cell lines.
After fixation, samples can be stored immediately at -80C or transported on dry ice and then stored at -80C. Current storage times are up to 6 months.
The first distribution of cells into the Reverse Transcription (RT) plate assigns a well-specific barcode that is used to identify the sample(s) during data analysis. Samples can be distributed across multiple wells for deeper sample profiling, or each well can contain an individual sample for maximum sample throughput.
ScaleBio provides a downstream data analysis pipeline, the ScaleBio Seq Suite, which is compatible with sequencing instrument output files. The pipeline will de-multiplex samples, generate a cell-gene matrix, as well as provide a data quality metrics summary to assess run performance.
ScaleBio RNA libraries have been demonstrated with MiSeq, NextSeq and NovaSeq instruments.

Want to learn more about our Single Cell RNA Sequencing Kit?
Reach out to our Team for more information or to request a quote.
Related products
